Focus on diagnostics
We process the data
Cloud-based genetic diagnostics to accelerate laboratory workflows and enhance patient care.
One turnkey solution for all clinical NGS applications
Solution
The varvis® genomics software is a complete solution for clinical diagnostics, supporting both short- and long-read sequencing. It is designed to streamline the entire NGS workflow, including raw data processing, genomic data management, and variant interpretation. The platform integrates automated analysis of SNVs, CNVs, STRs, and SVs demonstrating robust performance across diverse panel sizes, including whole genome sequencing
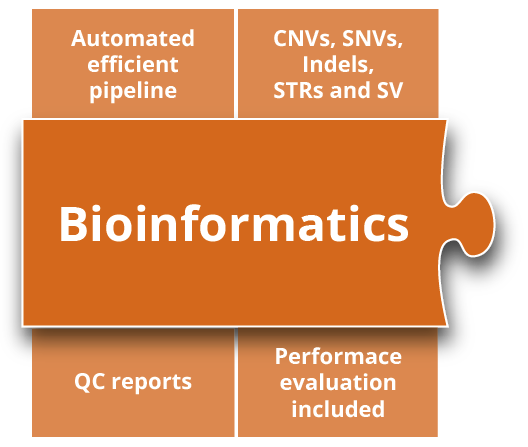
Bioinformatics
Truely hands-free.
Our automated bioinformatics pipeline processes raw short- and long-read sequencing data from bcl, fastq or bam files. On completion of the bioinformatics analyses, including alignment and variant calling of SNV, CNVs, STRs and SVs, all results are annotated and visualized in the varvis® software automatically. Performance test reports are provided as a service – updates are included.
varvis® genomics software
Made for use in clinical diagnostics
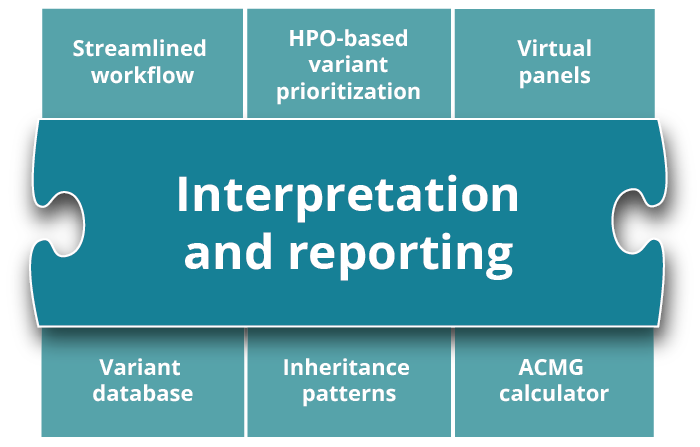
The varvis® software is a clinical decision support system (CDS) and allows you to review, filter, and classify genetic variants. In addition, it includes your own comprehensive variant database. It supports the clinical decision-making process and is the first genomics end-to-end software certified as IVDR Class C device.
Annotation
Always up to date
Our reference database, allexes®, provides the data for variant annotation to the varvis® software. The allexes® database does not only deliver the most recent versions of public databases, but also provides access to aggregated genomic reference data from all our users while continuing to be compliant with HIPAA and EU regulations.
Key benefits
Automated
quality control
Important quality metrics are monitored automatically for every single NGS sample, but also across batches over time. Don't waste a moment on manual QC.
Push
the button
Convenient filtering options such as inheritance filters and virtual panels allow you to filter from thousands of detected SNVs and CNVs. Within seconds.
Supreme expert support
Our dedicated team provides first-class support regarding workflow optimization, technical issues, training and documentation – even for the tricky cases. We are here to help!
Trust
your results
Simply sequence the appropriate reference sample to validate your workflow – we take care of the rest and provide you with a performance test report. Regular updates are included.
Use cases
Targeted
panels
Identify rare and causative SNVs and CNVs in a single process of proven performance. Replace conventional PCR-based methods.
Whole exome
sequencing
Do you need more than 15 minutes to interpret a whole exome? Accelerate WES interpretation by combining phenotype, family and inheritance information.
Long-read sequencing
Are you ready for long read sequencing in clinical diagnostics? Discover the dark genome and detect repeat expansions with confidence.
Carrier
screening
Even the most complex scenarios are well covered in a single carrier screening filter to solve your cases with just a few clicks.
See for yourself how the varvis® software can accelerate your laboratory workflows and increase your diagnostic yield.
Testimonials

University of Magdeburg

University of Leipzig

Synlab Zentrum für Humangenetik Mannheim

University of Göttingen

Gemeinschaftspraxis für Humangenetik & Genetische Labore Hamburg
Read more

Update to gnomAD v4.1: Key features
by Dr. Roberta Trunzo, August 27, 2024
The varvis® software now includes the latest version of the Genome Aggregation Database (gnomAD), providing updated annotations for whole exome and whole genome data. This article describes the enhancements and implications of the update to the new version 4.1, particularly for clinical diagnostics.

The varvis® Software: The first genomics end-to-end software certified as IVDR Class C device
by Dr. Ben Liesfeld, May 31, 2024
Genetic diagnostic laboratories now have access to the first complete genomics software solution which is certified as a Class C device under IVDR. This will significantly reduce the effort required for legally compliant documentation of in-house tests.
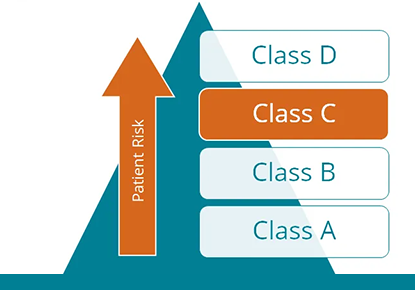
Q&A: The role of the IVDR conformity assessment in genetic diagnostics
by Dr. Ben Liesfeld & Dr. Sonja Strunz,
May 31, 2024
Now that the regulation on in-vitro diagnostic devices (IVDR) explicitly regulates in-house devices, or laboratory-developed tests (LDTs), in the European Union, the selection of properly CE-labeled devices is increasingly important to health institutions like genetic testing laboratories.'